
Photo: Greg Kahn, Howard Hughes Medical Institute
In its effort to correlate genomic structure with gene function, the 4D Nucleome Consortium (4DN), led by Job Dekker, PhD, at UMass Chan Medical School, has extensively mapped and analyzed the three-dimensional folding of the human genome in human embryonic stem cells and immortalized fibroblasts over time. The result is the most detailed view of the four-dimensional human genome available and the identification of more than 140,000 looping interactions between genes and long-range regulatory elements. The study was published in Nature.
Integrating data from more than a dozen benchmarked genomic and computational techniques, the 4DN has created a critical framework for future studies that will further scientific inquiry into the architecture of the genome and how structural variations are associated with gene function in cells.
“To understand how DNA turns linear genetic information in the form of nucleotides into biologically meaningful actions, it’s essential to map the physical organization of the genome in space relative to itself and other landmarks and bodies in the nucleus, as well as how this structure varies from cell to cell and state to state,” said Dr. Dekker, Howard Hughes Medical Investigator, the Joseph J. Byrne Chair in Biomedical Research and professor of systems biology. “The map produced in this study by the 4DN Consortium allows us to visualize how the genome is organized in both space and over time, and how it relates to genome function.”
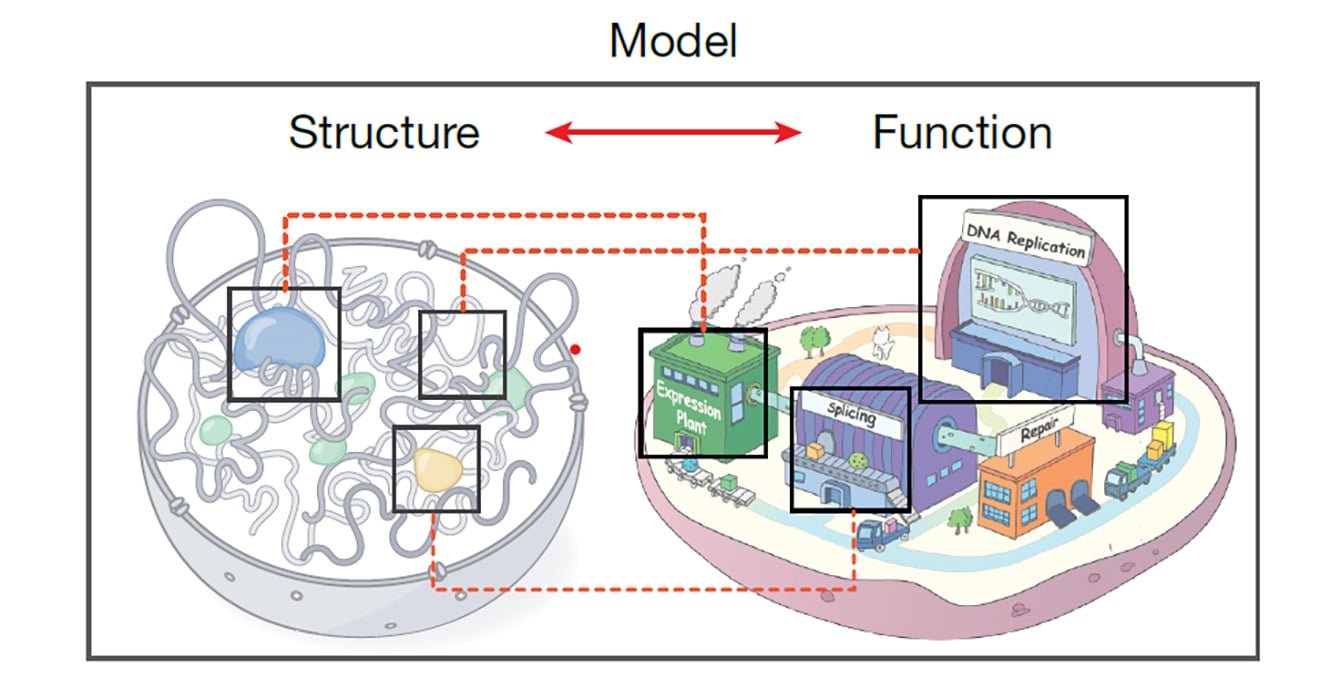
Illustration provided by the Dekker lab.
The human genome contains more than 20,000 protein-coding genes with an estimated millions of regulatory elements that influence those genes. Scientific studies have catalogued some of these components. However, the mechanisms by which these regulatory elements act on specific genes, especially over long distances of up to hundreds of kilobases and sometime megabases, are still poorly understood.
How the genome is folded, organized and structured in space greatly influences what regulatory elements and genes are in proximity to each other, as well as their location in relation to other molecular bodies in the nucleus. This, in part, explains how elements that are spaced widely apart along the linear genome can interact in three-dimensional space. Like cooked spaghetti gathered in a bowl, folding and looping brings distant elements into proximity to each other so they can interact. What’s more, the structure of the genome can vary from cell to cell and state to state, such as during cell division or when a cell undergoes stress, further influencing gene function.
“After determining the complete DNA sequence of the human genome and the subsequent mapping of most genes and identification of potential regulatory elements, we are now in a position that can be considered the third phase of the human genome project: its spatial organization,” said Dekker.
Since its founding in 2015, a major focus of the 4DN has been the development and benchmarking of complementary experimental approaches for measuring the genome in three-dimensional space, as well as over time.
The latest study integrates work from more than three dozen labs across eight countries. It includes data derived using experimental methodologies, such as Hi-C, micro-C, SPRITE, ChiA-Pet, PLAC-seq and GAM, as well as numerous computational tools and modeling approaches for analyzing and interpreting data. The study outlines how each experimental technique contributes unique and overlapping information about the nature of genomic folding. It elucidates connections between chromosome folding and looping, nuclear positioning and proximity to nuclear bodies, as well as cell-to-cell variation in organization and genomic processes such as transcription and replication.
The culmination of this work is a scientific user’s guide in the form of a table and decision tree that details the strengths of various assays, how each of those methods contribute fundamental knowledge to the elucidation of the three-dimensional genome, and, more importantly, which methods to use for specific research questions.
By integrating and combining different datasets, 4DN has, among other findings, compiled an extensive catalogue of 140,000 looping interactions between specific elements that no single methodology could have detected on its own. The program also used integrated models to begin assigning dynamic genomic functions comprised of multiple genes and regulatory elements, such as replication and transcription, into a three-dimensional context.
Further, the group showed how the data can be used to train deep learning models and AI that can then be used to screen DNA sequences to uncover genome folding mechanisms and their association with gene function.
“This is the most detailed view of the living physical genome as it exists inside of cells,” said Dekker. “It’s the foundation for the deep exploration of structure and function of the genome.”
The next phase of the 4DN project is to integrate genomic datasets with new, advanced imaging data and analyze nuclear changes during development and disease.
Oliver J. Rando, PhD, the Endowed Chair in Biochemistry and Molecular Biotechnology II and professor of biochemistry & molecular biotechnology at UMass Chan, is a co-author of the study.
This work has been collectively funded by grants from the National Institutes of Health Common Fund (The 4D Nucleome Project). A collection of 4DN project studies is available on the Nature web site.

